Tutorial
This page explains how to use the
RPISeq.
The contents for the tutorial are:
- RPISeq information
- Example: Basic function
- Example: Batch submission
- Example: Query against the database
RPISeq Information
Background:
RNA-protein interactions (RPIs) play important roles in a wide variety of cellular processes. High throughput experiments to identify RNA-protein interactions are beginning to provide valuable information about the complexity of RNA-protein interaction networks, but are expensive and time consuming. RPISeq was developed to address the need for reliable computational methods for predicting RNA-protein interactions.
RPISeq is a family of machine learning classifiers for predicting RNA-protein interactions using only sequence information.
- RPISeq predictions are based on Random Forest (RF) or Support Vector Machine (SVM) classifiers trained and tested on 2 non-redundant benchmark datasets of RNA-protein interactions, RPI2241 and RPI369, extracted from PRIDB, a comprehensive database of RNA-protein complexes extracted from the PDB.
- The RPISeq server can:
- predict the probability that a specific protein and RNA interact
(user provides one protein and one RNA sequence)
- for a given protein sequence, identify its most likely RNA binding partner from user-provided list
(user provides one protein sequence and a list of up to 100 RNA sequences)
- for a given RNA sequence, identify its most likely protein binding partner from a user-provided list
(user provides one RNA sequence and a list of up to 100 protein sequences)
- use a given protein sequence to query a database of known RNA-protein interactions (RPIntDB)
(user provides one protein sequence)
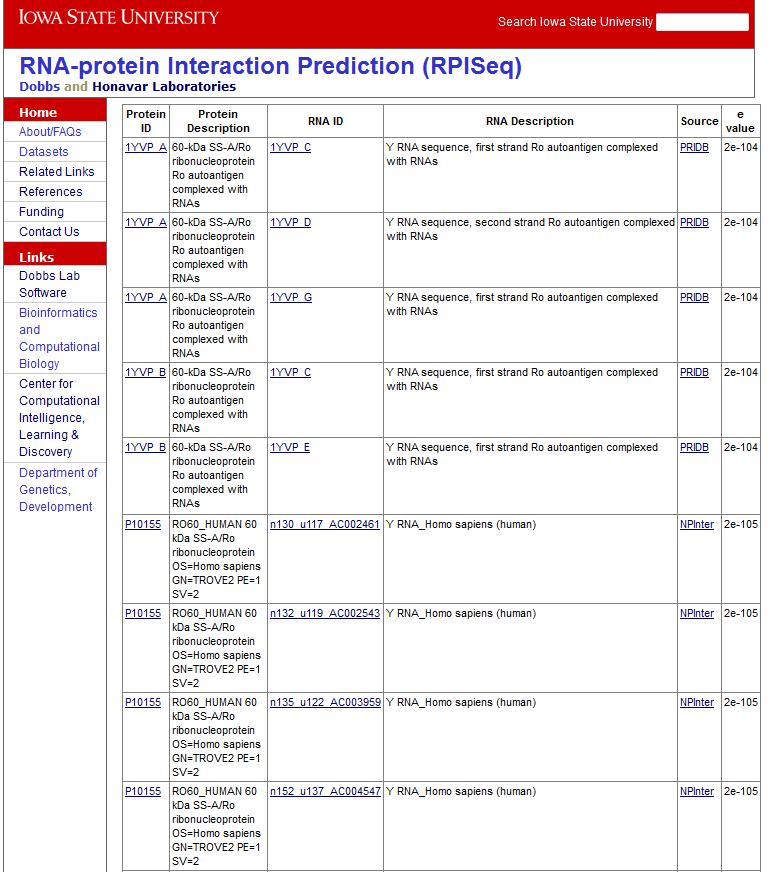
- RPIntDB - a database of known RNA-Protein interactions
RPIntDB contains a total of 30,056 RNA-Protein interactions (5694 unique RNA chains and 1702 unique protein chains). The datasets can be downloaded from here.
Two types of experimentally validated RNA-Protein interactions are represented in RPIntDB:
- PRIDB - RNA-protein interactions extracted from PRIDB, a database of RNA protein interfaces.
These include RNA and protein chains identified as interacting in experimentally-determined (X-ray and NMR) structures of RNA-protein complexes derived from the PDB; for details, please see: Lewis et al., 2011 Nucleic Acids Research
- NPInter - RNA-protein interactions extracted from NPInter, a database of non-coding RNA (ncRNA) protein interactions. These include RNA and protein chains identified as interacting; for details, please see: Wu et al., 2005 Nucleic Acids Research
- High-throughput experimental data - These interactions are extracted from available literature. Links to the corresponding publications are provided in the 'Source' column.
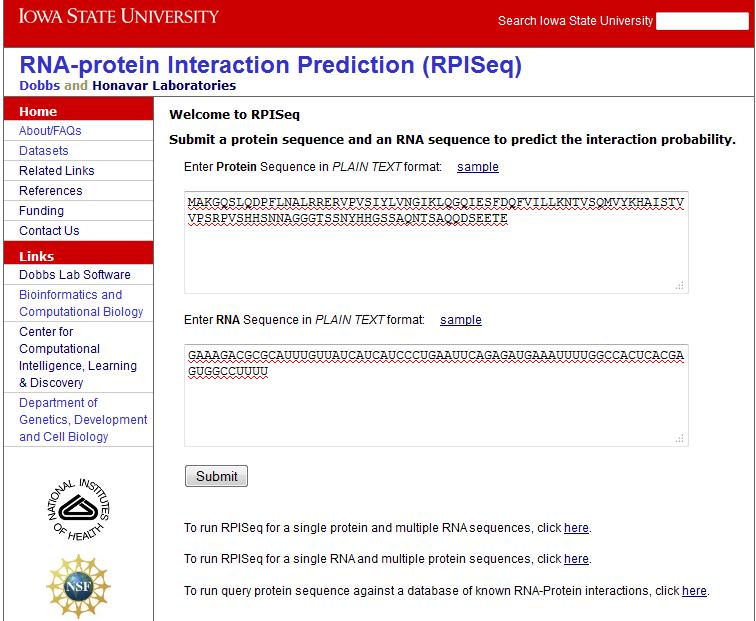
Example: Basic function
Enter a protein sequence and a RNA sequence in the text boxes provided and click 'Submit'. Figure 1 illustrates the basic function with sample protein and RNA sequences filled in:
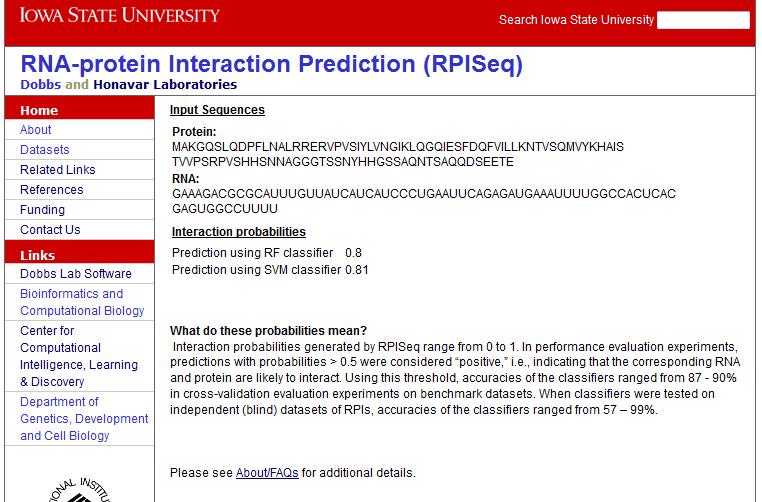
The output displays the sequences submitted and the probabilities ofinteraction predicted using Random Forest and SVM classifiers. Figure 2 shows the output using the sample protein and RNA sequences.
Example: Batch submission
If you have multiple RNA or protein sequences, upload the file containing sequences in FASTA format. Click Submit. Figure 3 shows the batch submission with a sample protein and a file of RNA sequences in FASTA format:
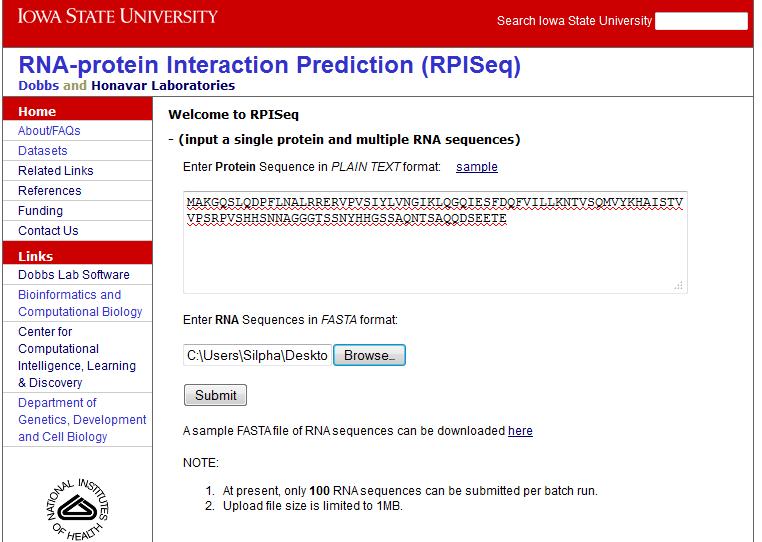
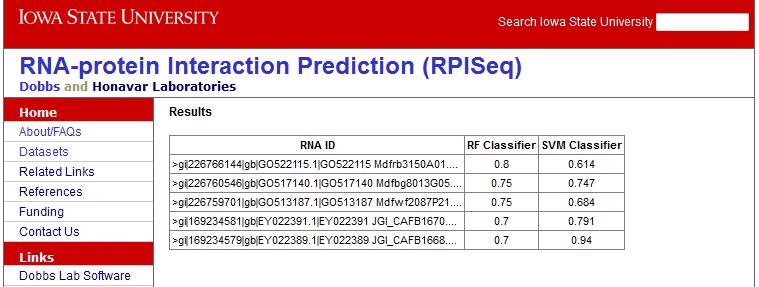
Figure 4 with sample output is shown below:
Example: Query against a database
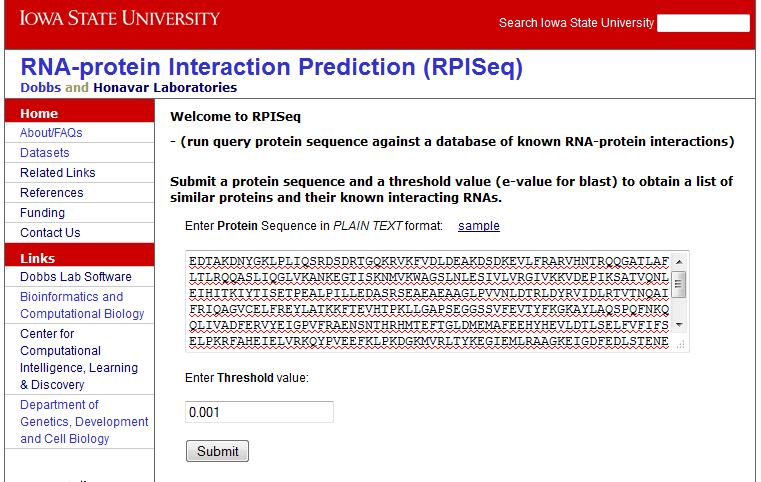
Input a single protein sequence in plain text format. Click 'Submit'. Figure 5 shows the query page with sample protein data:
The output page displays the protein hits and interacting RNAs and reference for the interaction. Links to SwissProt, PRIDB, NPInter and other available literature from where these interactions are obtained are also provided.